Machine learning model for fast prediction of the natural frequencies of protein molecules†
Abstract
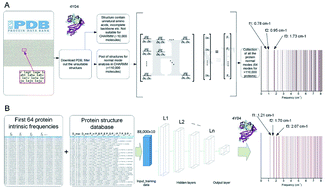
Natural vibrations and resonances are intrinsic features of protein structures and enable differentiation of one structure from another. These nanoscale features are important to help to understand the dynamics of a protein molecule and identify the effects of small sequence or other geometric alterations that may not cause significant visible structural changes, such as point mutations associated with disease or drug design. Although normal mode analysis provides a powerful way to accurately extract the natural frequencies of a protein, it must meet several critical conditions, including availability of high-resolution structures, availability of good chemical force fields and memory-intensive large-scale computing resources. Here, we study the natural frequency of over 100 000 known protein molecular structures from the Protein Data Bank and use this dataset to carefully investigate the correlation between their structural features and these natural frequencies by using a machine learning model composed of a Feedforward Neural Network made of four hidden layers that predicts the natural frequencies in excellent agreement with full-atomistic normal mode calculations, but is significantly more computationally efficient. In addition to the computational advance, we demonstrate that this model can be used to directly obtain the natural frequencies by merely using five structural features of protein molecules as predictor variables, including the largest and smallest diameter, and the ratio of amino acid residues with alpha-helix, beta strand and 3–10 helix domains. These structural features can be either experimentally or computationally obtained, and do not require a full-atomistic model of a protein of interest. This method is helpful in predicting the absorption and resonance functions of an unknown protein molecule without solving its full atomic structure.
- This article is part of the themed collection: New Insights into Biomolecular Systems from Large-Scale Simulations


 Please wait while we load your content...
Please wait while we load your content...