A core–shell CdTe quantum dots molecularly imprinted polymer for recognizing and detecting p-nitrophenol based on computer simulation†
Abstract
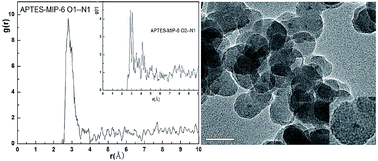
In this study, molecularly imprinted technology, combined with fluorescence measurement and computer simulation, was used to detect contaminant p-nitrophenol. Seven molecular dynamics simulations of molecular imprinting prepolymerization systems were performed to optimize the imprinting shell. Results indicated that the system with a p-nitrophenol (4-NP): 3-aminopropyltriethoxysilane (APTES): tetraethylorthosilicate (TEOS) mole ratio of 8 : 8 : 12 led to the best stable template–functional monomer clusters and the hybrid SiO2 layer with CdS-like clusters on the surface of CdTe was synthesized by a simple reflux procedure. The prepared imprinted materials (CdTe@SiO2-MIPs) on the surface of silica contained CdTe nanoparticles (CdTe@SiO2) by surface imprinting and Stöber method polymerization were characterized by transmission electron microscopy, fluorescence spectroscopy, Fourier-transform infrared spectroscopy, ultraviolet–visible analysis and X-ray diffraction. A linear relationship between relative fluorescence intensity and the concentration of 4-NP was obtained with a limit of detection of 0.08 μmol L−1 and the imprinting factor (IF) was 2.23 which indicated that special binding sites with binding property to p-nitrophenol were created on the surface of the CdTe@SiO2-MIPs materials. Ultimately, the feasibility of the fluorescent materials was successfully evaluated through the analysis of 4-NP in tap water and lake water. The recoveries were above 97.3%.

 Please wait while we load your content...
Please wait while we load your content...