Structure–activity relationship study for design of highly active covalent peroxidase-mimicking DNAzyme†
Abstract
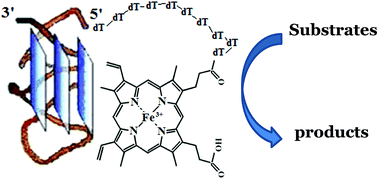
We synthesized a series of conjugates of hemin and its aptamer EAD2, named covalent peroxidase-mimicking DNAzymes (PMDNAzymes), varying the length, rigidity and 5′-/3′-position of a linker between the oligonucleotide and hemin. Systemic structure–activity relationship study of these PMDNAzymes showed that covalent PMDNAzyme with hemin bound to the 5′-end of EAD2 via T10 spacer (PMDNAzyme(T10)) demonstrated the highest activity in luminol oxidation assay. Its activity was significantly higher in comparison to the non-covalent complex of hemin and aptamer EAD2 (non-covalent PMDNAzyme). Comparison of the detection limit values for the PMDNAzyme(T10) in the reactions of oxidation of luminol and ABTS, which were equal to 0.2 and 1.6 pM, respectively, showed that the chemiluminescent method of PMDNAzyme(T10) detection is preferred over the colorimetric one. Similarity of the detection limit values for the PMDNAzyme(T10) and horseradish peroxidase, whose activity was measured in an enhanced chemiluminescence reaction (0.25 pM), opens up very promising perspectives for the development of highly sensitive PMDNAzyme(T10)-based assays and devices.

 Please wait while we load your content...
Please wait while we load your content...