In silico screening of molecular imprinting prepolymerization systems: oseltamivir selective polymers through full-system molecular dynamics-based studies†
Abstract
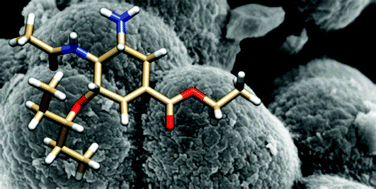
All-component molecular dynamics studies were used to probe a library of oseltamivir molecularly imprinted polymer prepolymerization mixtures. Polymers included one of five functional monomers (acrylamide, hydroxyethylmethacrylate, methacrylic acid, 2-(triflouromethyl)acrylic acid, 4-vinylpyridine) and one of three porogens (acetonitrile, chloroform, methanol) combined with the crosslinking agent ethylene glycol dimethacrylate and initiator 2,2′-azobis(2-methylpropionitrile). Polymers were characterized by nitrogen gas sorption measurements and SEM, and affinity studies performed using radioligand binding in various media. In agreement with the predictions made from the simulations, polymers prepared in acetonitrile using either methacrylic or trifluoromethacrylic acid demonstrated the highest affinities for oseltamivir. Further, the ensemble of interactions observed in the methanol system provided an explanation for the morphology of polymers prepared in this solvent. The materials developed here offer potential for use in solid-phase extraction or for catalysis. The results illustrate the strength of this in silico strategy as a potential prognostic tool in molecularly imprinted polymer design.
 Please wait while we load your content...
Please wait while we load your content...