Preparation and conformational analysis of polyproline tri-helix macrocycle nanoscaffolds of varied sizes†
Abstract
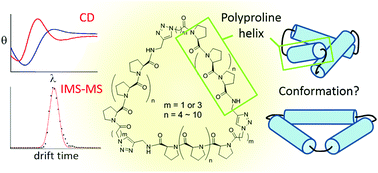
Ligand patterns at the nanoscale are essential in modulating biological recognition and signaling through binding to receptor oligomers. Biocompatible nanoscaffolds that allow precise control of multiple ligand presentation would be of great use in manipulating cellular processes and understanding membrane receptor biology. We have previously developed tri-helix and tetra-helix macrocycle scaffolds based on the Pro9 peptide helix to control ligand arrangements that can selectively target receptor oligomers. A better understanding of the structure of these macromolecules would significantly reduce the difficulty in designing matching ligand positions for target receptors. In this work, we expand the arsenal of ligand patterns by preparing polyproline tri-helix macrocycle scaffolds of different sizes. These synthetic nanoscaffolds composed of peptide helices ranging from Pro6 to Pro12 also allowed us to systematically investigate their properties. With a combination of circular dichroism spectroscopy and ion mobility spectrometry–mass spectrometry (IMS-MS), the measurement for varied sizes of these scaffolds indicated the connecting dihedral angle between both ends of the helix affects the strain in the cyclic scaffold. The experimental collision cross section obtained from IMS-MS favors a propeller model for the helix arrangements. The results not only contribute conformational insights for the polyproline tri-helix system, but also provide precious information for the future design and synthesis of cyclic nanostructures based on peptide helices.


 Please wait while we load your content...
Please wait while we load your content...