Live cell single molecule tracking and localization microscopy of bioorthogonally labeled plasma membrane proteins†
Abstract
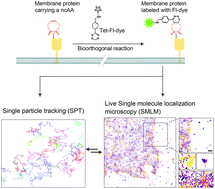
Tracking the localization and mobility of individual proteins in live cells is key for understanding how they mediate their function. Such information can be obtained from single molecule imaging techniques including as Single Particle Tracking (SPT) and Single Molecule Localization Microscopy (SMLM). Genetic code expansion (GCE) combined with bioorthogonal chemistry offers an elegant approach for direct labeling of proteins with fluorescent dyes, holding great potential for improving protein labeling in single molecule applications. Here we calibrated conditions for performing SPT and live-SMLM of bioorthogonally labeled plasma membrane proteins in live mammalian cells. Using SPT, the diffusion of bioorthogonally labeled EGF receptor and the prototypical Shaker voltage-activated potassium channel (Kv) was measured and characterized. Applying live-SMLM to bioorthogonally labeled Shaker Kv channels enabled visualizing the plasma membrane distribution of the channel over time with ∼30 nm accuracy. Finally, by competitive labeling with two Fl-dyes, SPT and live-SMLM were performed in a single cell and both the density and dynamics of the EGF receptor were measured at single molecule resolution in subregions of the cell. We conclude that GCE and bioorthogonal chemistry is a highly suitable, flexible approach for protein labeling in quantitative single molecule applications that outperforms current protein live-cell labeling approaches.


 Please wait while we load your content...
Please wait while we load your content...