Blocking probe as a potential tool for detection of single nucleotide DNA mutations: design and performance†
Abstract
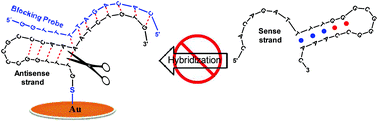
Developing strategies to detect single nucleotide DNA mutations associated with treatment decisions in cancer patients from liquid biopsies is a rapidly emerging area of personalized medicine that requires high specificity. Here we report how to design an easy enzyme-free approach that could create a platform for detection of L858R mutation of EGFR that is a predictive biomarker of tyrosine kinase treatment in many cancers. This approach includes the addition of blocking probes with the antisense ssDNA at different blocking positions and different concentrations such as to avoid re-annealing with the respective sense ssDNA. The successful blocking strategy was corroborated by fluorescence spectroscopy in solution using two distinct FRET pairs and quartz crystal microbalance with dissipation (QCM-D) measurements under comparable experimental conditions, as the hybridization rate-limiting step in both methods is the nucleation process. The efficiency of hybridization of each blocking probe was strongly dependent on its position particularly when the analyte possesses a secondary hairpin-structure. We tested the performance of blocking probes in combination with gold nanoparticles; the obtained results were in agreement with those of QCM-D. These findings could facilitate the development of better biosensors, especially those using probes containing secondary structure.


 Please wait while we load your content...
Please wait while we load your content...