Computational genomic identification and functional reconstitution of plant natural product biosynthetic pathways
Abstract
Covering: 2003 to 2016
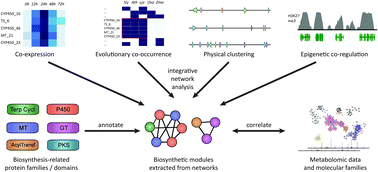
The last decade has seen the first major discoveries regarding the genomic basis of plant natural product biosynthetic pathways. Four key computationally driven strategies have been developed to identify such pathways, which make use of physical clustering, co-expression, evolutionary co-occurrence and epigenomic co-regulation of the genes involved in producing a plant natural product. Here, we discuss how these approaches can be used for the discovery of plant biosynthetic pathways encoded by both chromosomally clustered and non-clustered genes. Additionally, we will discuss opportunities to prioritize plant gene clusters for experimental characterization, and end with a forward-looking perspective on how synthetic biology technologies will allow effective functional reconstitution of candidate pathways using a variety of genetic systems.
- This article is part of the themed collection: Synthetic Biology and Bioinformatics

 Please wait while we load your content...
Please wait while we load your content...