Matched triplicate design sets in the optimisation of glucokinase activators – maximising medicinal chemistry information content†
Abstract
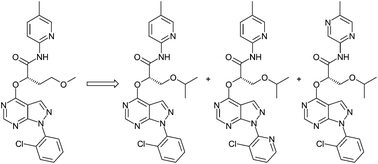
Successful lead optimisation requires the identification of the best compound within the chemical space explored during an optimisation campaign. This can be a costly and inefficient process leading to the synthesis of many sub-optimal compounds. In this paper, a method for carrying out this exercise more effectively is outlined. This relies on the generation of robust datasets on which to build predictive models in a paradigm termed “matched triplicate design sets”. The practical implementation of this approach is exemplified in the optimisation of a new series of glucokinase activators.

 Please wait while we load your content...
Please wait while we load your content...