Identification of human flap endonuclease 1 (FEN1) inhibitors using a machine learning based consensus virtual screening†
Abstract
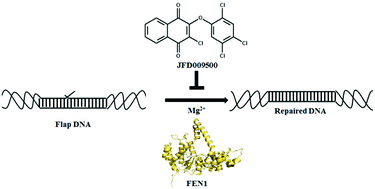
Human Flap endonuclease1 (FEN1) is an enzyme that is indispensable for DNA replication and repair processes and inhibition of its Flap cleavage activity results in increased cellular sensitivity to DNA damaging agents (cisplatin, temozolomide, MMS, etc.), with the potential to improve cancer prognosis. Reports of the high expression levels of FEN1 in several cancer cells support the idea that FEN1 inhibitors may target cancer cells with minimum side effects to normal cells. In this study, we used large publicly available, high-throughput screening data of small molecule compounds targeted against FEN1. Two machine learning algorithms, Support Vector Machine (SVM) and Random Forest (RF), were utilized to generate four classification models from huge PubChem bioassay data containing probable FEN1 inhibitors and non-inhibitors. We also investigated the influence of randomly selected Zinc-database compounds as negative data on the outcome of classification modelling. The results show that the SVM model with inactive compounds was superior to RF with Matthews's correlation coefficient (MCC) of 0.67 for the test set. A Maybridge database containing approximately 53 000 compounds was screened and top ranking 5 compounds were selected for enzyme and cell-based in vitro screening. The compound JFD00950 was identified as a novel FEN1 inhibitor with in vitro inhibition of flap cleavage activity as well as cytotoxic activity against a colon cancer cell line, DLD-1.


 Please wait while we load your content...
Please wait while we load your content...