plantMirP: an efficient computational program for the prediction of plant pre-miRNA by incorporating knowledge-based energy features†
Abstract
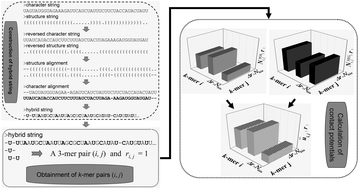
MicroRNAs are a predominant type of small non-coding RNAs approximately 21 nucleotides in length that play an essential role at the post-transcriptional level by either RNA degradation, translational repression or both through an RNA-induced silencing complex. Identification of these molecules can aid the dissecting of their regulatory functions. The secondary structures of plant pre-miRNAs are much more complex than those of animal pre-miRNAs. In contrast to prediction tools for animal pre-miRNAs, much less effort has been contributed to plant pre-miRNAs. In this study, a set of novel knowledge-based energy features that has very high discriminatory power is proposed and incorporated with the existing features for specifically distinguishing the hairpins of real/pseudo plant pre-miRNAs. A promising performance area under a receiver operating characteristic curve of 0.9444 indicates that 5 knowledge-based energy features have very high discriminatory power. The 10-fold cross-validation result demonstrates that plantMirP with full features has a promising sensitivity of 92.61% and a specificity of 98.88%. Based on various different datasets, it was found that plantMirP has a higher prediction performance by comparison with miPlantPreMat, PlantMiRNAPred, triplet-SVM, and microPred. Meanwhile, plantMirP can greatly balance sensitivity and specificity for real/pseudo plant pre-miRNAs. Taken together, we developed a promising SVM-based program, plantMirP, for predicting plant pre-miRNAs by incorporating knowledge-based energy features. This study shows it to be a valuable tool for miRNA-related studies.

 Please wait while we load your content...
Please wait while we load your content...