Estimation of delays in generalized asynchronous Boolean networks†
Abstract
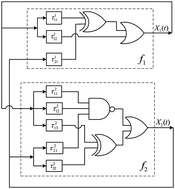
A new generalized asynchronous Boolean network (GABN) model has been proposed in this paper. This continuous-time discrete-state model captures the biological reality of cellular dynamics without compromising the computational efficiency of the Boolean framework. The GABN synthesis procedure is based on the prior knowledge of the logical structure of the regulatory network, and the experimental transcriptional parameters. The novelty of the proposed methodology lies in considering different delays associated with the activation and deactivation of a particular protein (especially the transcription factors). A few illustrative examples of some well-studied network motifs have been provided to explore the scope of using the GABN model for larger networks. The GABN model of the p53-signaling pathway in response to γ-irradiation has also been simulated in the current paper to provide an indirect validation of the proposed schema.

 Please wait while we load your content...
Please wait while we load your content...