MarRA, SoxSR, and Rob encode a signal dependent regulatory network in Escherichia coli†
Abstract
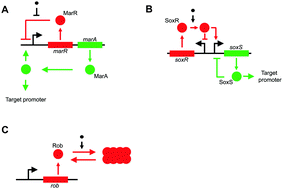
When exposed to low concentrations of toxic chemicals, bacteria modulate the expression of a number of cellular processes. Typically, these processes include those related to porin production, dismutases, and metabolic fluxes. In Escherichia coli (E. coli), the expression of these systems is largely controlled by three homologous transcriptional regulators: MarA, SoxS, and Rob. Each of the three regulators responds to distinct chemical signals (salicylate for MarA; paraquat for SoxS; and bipyridyl for Rob) and controls the expression of an overlapping set of downstream targets. In addition, the three systems autoregulate their own expression, and cross-regulate each other's expression. Specifically, MarA is known to activate SoxS expression, and Rob is known to activate MarA expression. In addition, a number of conflicting regulatory interactions are known to exist between the three loci. Thus, the three systems encode a complex regulatory topology with multiple feedback loops, the precise nature of whose interactions or their significance in cellular physiology is not well understood currently. In this work, we focus on understanding the details of this crosstalk between the Mar–Sox–Rob systems in E. coli, and the resulting control and dynamics of the expression of cellular processes by studying gene expression at the population level and at single-cell resolution in wild type and mutants. Our results indicate that the regulatory architecture between MarA, SoxS, and Rob is dependent on the signal (inducer) present in the environment. The regulators, in response to an inducer, form a Feed Forward Loop (FFL), which leads to faster and stronger induction of target genes in the cell, consequently resulting in better cellular growth. Through the FFL, the cell is able to integrate qualitatively different signals in the network, and consequently, control cellular physiology. In addition, we present two intriguing dynamic features of the Mar–Sox–Rob regulon. First, in the presence of salicylate, the activation of target genes via MarA and Rob, at single-cell resolution, is qualitatively different. Second, we report the synergistic activation of target and Mar/Sox systems in the presence of both salicylate and paraquat. These results strongly indicate that there exists a complex control of gene regulation in the Mar–Sox–Rob regulon. Mechanistic details of this control are likely quite complex, and may involve additional regulators.

 Please wait while we load your content...
Please wait while we load your content...