Analyses of methyltransferases across the pathogenicity spectrum of different mycobacterial species point to an extremophile connection†
Abstract
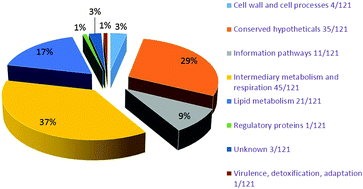
Tuberculosis is a devastating disease, taking one human life every 20 seconds globally. We hypothesize that professional pathogens such as M.tb have acquired specific features that might assist in causing infection, persistence and transmissible pathology in their host. We have identified 121 methyltransferases (MTases) in the M.tb proteome, which use a variety of substrates – DNA, RNA, protein, intermediates of mycolic acid biosynthesis and other fatty acids – that are involved in cellular maintenance within the host. A comparative analysis of the proteome of the virulent strain H37Rv and the avirulent strain H37Ra identified 3 MTases, which displayed significant variations in terms of N-terminal extension/deletion and point mutations, possibly impacting various physicochemical properties. The cross-proteomic comparison of MTases of M.tb H37Rv with 15 different Mycobacterium species revealed the acquisition of novel MTases in a MTB complex as a function of evolution. Phylogenetic analysis revealed that these newly acquired MTases showed common roots with certain extremophiles such as halophilic and acidophilic organisms. Our results establish an evolutionary relationship of M.tb with halotolerant organisms and also the role of MTases of M.tb in withstanding the host osmotic stress, thereby pointing to their likely role in pathogenesis, virulence and niche adaptation.

 Please wait while we load your content...
Please wait while we load your content...