In silico research to assist the investigation of carboxamide derivatives as potent TRPV1 antagonists†
Abstract
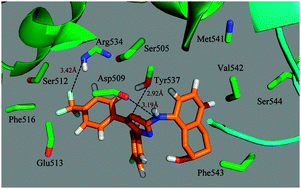
The transient receptor potential vanilloid type 1 (TRPV1), a non-selective cation channel, is known for its essential role in the pathogenesis of various pain conditions such as nerve damage induced hyperalgesia, diabetic neuropathy and cancer pain. Therefore, TRPV1 is considered as a promising target for the development of new anti-inflammatory and analgesic drugs. In the present study, a theoretical study on the functionalities of the molecular interactions between 236 active ligands and TRPV1 was carried out, using three-dimensional quantitative structure–activity relationships (3D-QSAR), molecular docking and molecular dynamics (MD) simulation approaches. The ligand-based CoMSIA model (obtained by use of a random division method for splitting the training and test sets) exhibits optimum predictivity (Q2 = 0.522, Rncv2 = 0.935, Rpred2 = 0.839). The results show that the models are useful tools for the prediction of the test sets as well as newly designed structures against TRPV1 activity. In addition, to verify the rationality of the random division method for splitting the dataset, we also used a self-organizing map (SOM) division approach for establishing the QSAR models. Interestingly, the obtained optimal CoMSIA model based on the SOM division exhibits almost the same proper statistical results (Q2 = 0.521, Rncv2 = 0.929, Rpred2 = 0.829) as the random division-derived model, proving the reasonability of both division methods for building the models. The contour plots of molecular fields along with docking and MD simulation have identified several key structural requirements responsible for the activity. The present work provides extremely useful guidelines for future structural modifications of this class of compounds towards the development of superior TRPV1 antagonists. The new computational insights presented in this study are expected to be valuable for the guideline and development of new potent TRPV1 antagonists.

 Please wait while we load your content...
Please wait while we load your content...