Experimental and computational studies of fatty acid distribution networks
Abstract
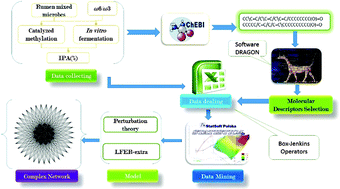
Unbalanced uptake of Omega 6/Omega 3 (ω-6/ω-3) ratios could increase chronic disease occurrences, such as inflammation, atherosclerosis, or tumor proliferation, and methylation methods for measuring the ruminal microbiome fatty acid (FA) composition/distribution play a vital role in discovering the contribution of food components to ruminant products (e.g., meat and milk) when pursuing a healthy diet. Hansch's models based on Linear Free Energy Relationships (LFERs) using physicochemical parameters, such as partition coefficients, molar refractivity, and polarizability, as input variables (Vk) are advocated. In this work, a new combined experimental and theoretical strategy was proposed to study the effect of ω-6/ω-3 ratios, FA chemical structure, and other factors over FA distribution networks in the ruminal microbiome. In step 1, experiments were carried out to measure long chain fatty acid (LCFA) profiles in the rumen microbiome (bacterial and protozoan), and volatile fatty acids (VFAs) in fermentation media. In step 2, the proportions and physicochemical parameter values of LCFAs and VFAs were calculated under different boundary conditions (cj) like c1 = acid and/or base methylation treatments, c2 = with/without fermentation, c3 = FA distribution phase (media, bacterial, or protozoan microbiome), etc. In step 3, Perturbation Theory (PT) and LFER ideas were combined to develop a PT-LFER model of a FA distribution network using physicochemical parameters (Vk), the corresponding Box–Jenkins (ΔVkj) and PT operators (ΔΔVkj) in statistical analysis. The best PT-LFER model found predicted the effects of perturbations over the FA distribution network with sensitivity, specificity, and accuracy > 80% for 407 655 cases in training + external validation series. In step 4, alternative PT-LFER and PT-NLFER models were tested for training Linear and Non-Linear Artificial Neural Networks (ANNs). PT-NLFER models based on ANNs presented better performance but are more complicated than the PT-LFER model. Last, in step 5, the PT-LFER model based on LDA was used to reconstruct the complex networks of perturbations in the FA distribution and compared the giant components of the observed and predicted networks with random Erdős–Rényi network models. In short, our new PT-LFER model is a useful tool for predicting a distribution network in terms of specific fatty acid distribution.

 Please wait while we load your content...
Please wait while we load your content...