A microscopic insight from conformational thermodynamics to functional ligand binding in proteins†
Abstract
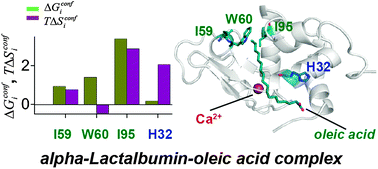
We show that the thermodynamics of metal ion-induced conformational changes aid to understand the functions of protein complexes. This is illustrated in the case of a metalloprotein, alpha-lactalbumin (aLA), a divalent metal ion binding protein. We use the histograms of dihedral angles of the protein, generated from all-atom molecular dynamics simulations, to calculate conformational thermodynamics. The thermodynamically destabilized and disordered residues in different conformational states of a protein are proposed to serve as binding sites for ligands. This is tested for β-1,4-galactosyltransferase (β4GalT) binding to the Ca2+–aLA complex, in which the binding residues are known. Among the binding residues, the C-terminal residues like aspartate (D) 116, glutamine (Q) 117, tryptophan (W) 118 and leucine (L) 119 are destabilized and disordered and can dock β4GalT onto Ca2+–aLA. No such thermodynamically favourable binding residues can be identified in the case of the Mg2+–aLA complex. We apply similar analysis to oleic acid binding and predict that the Ca2+–aLA complex can bind to oleic acid through the basic histidine (H) 32 of the A2 helix and the hydrophobic residues, namely, isoleucine (I) 59, W60 and I95, of the interfacial cleft. However, the number of destabilized and disordered residues in Mg2+–aLA are few, and hence, the oleic acid binding to Mg2+-bound aLA is less stable than that to the Ca2+–aLA complex. Our analysis can be generalized to understand the functionality of other ligand bound proteins.

 Please wait while we load your content...
Please wait while we load your content...