MCentridFS: a tool for identifying module biomarkers for multi-phenotypes from high-throughput data†
Abstract
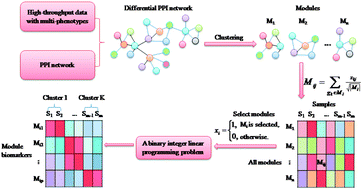
Systematically identifying biomarkers, in particular, network biomarkers, from high-throughput data is an important and challenging task, and many methods for two-class comparison have been developed to exploit information of high-throughput data. However, as the high-throughput data with multi-phenotypes are available, there is a great need to develop effective multi-classification models. In this study, we proposed a novel approach, called MCentridFS (Multi-class Centroid Feature Selection), to systematically identify responsive modules or network biomarkers for classifying multi-phenotypes from high-throughput data. MCentridFS formulated the multi-classification model by network modules as a binary integer linear programming problem, which can be solved efficiently and effectively in an accurate manner. The approach is evaluated with respect to two diseases, i.e., multi-stages HCV-induced dysplasia and hepatocellular carcinoma and multi-tissues breast cancer, both of which demonstrated the high classification rate and the cross-validation rate of the approach. The computational results of the five-fold cross-validation of the two data show that MCentridFS outperforms the state-of-the-art multi-classification methods. We further verified the effectiveness of MCentridFS to characterize the multi-phenotype processes using module biomarkers by two independent datasets. In addition, functional enrichment analysis revealed that the identified network modules are strongly related to the corresponding biological processes and pathways. All these results suggest that it can serve as a useful tool for module biomarker detection in multiple biological processes or multi-classification problems by exploring both big biological data and network information. The Matlab code for MCentridFS is freely available from http://www.sysbio.ac.cn/cb/chenlab/images/MCentridFS.rar.

 Please wait while we load your content...
Please wait while we load your content...