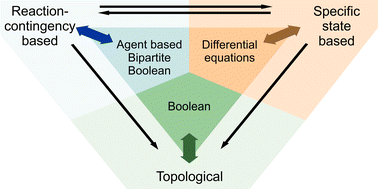
One of the first steps towards holistic understanding of cellular networks is the integration of the available information in a human and machine readable format. This network reconstruction process is well established for metabolic networks, and numerous genome wide metabolic reconstructions are already available. Extending these strategies to signalling networks has proven difficult, primarily due to the combinatorial nature of regulatory modifications. The combinatorial nature of possible protein–protein interactions and post translational modifications affects both network size and the correspondence between the reconstructed network and the underlying empirical data. Here, we discuss different approaches to reconstruction of signal transduction networks. We divide the current approaches into topological, specific state based and reaction-contingency based, and discuss their different information content and scalability. The discussion focusses on graphical formats but the points are in general applicable also to mathematical models and databases. While the formats have complementary strengths especially for small networks, reaction-contingency based formats have a number of advantages in the light of global network reconstruction. In particular, they minimise the need for assumptions, maximise the congruence with empirical data, and scale efficiently with network size.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...