Microfluidic auto-alignment of protein patterns for dissecting multi-receptor crosstalk in platelets
Abstract
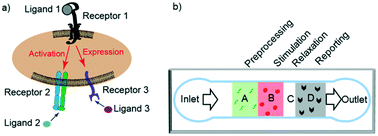
Cell adhesion plays a critical role in many cellular functions, such as the hemostatic/thrombotic process, inflammatory reactions, and adaptive immune response. Many cell adhesion processes involve crosstalk between multiple ligand–receptor systems through intracellular signaling. To elucidate such crosstalk requires analysis of the synergistic or antagonistic effects of binding and signalling of multi-receptor species. Current techniques for these analyses, e.g., atomic force microscopy (AFM) and biomembrane force probe (BFP) assays, are either labor-intensive, low-throughput, or limited in the types of ligands they can interrogate. Circumventing these limitations requires a technique for manipulating ligand interactions with and measuring the functional response of a population of cells. In this work, we have developed a microfluidic platform for studying the binding and signaling of multi-receptor species by separating their actions in space and time. The platform directs cells through a single channel and uses sequentially presented ligands for pre-processing and stimulating cells, followed by reporting of cell activation states and functional consequences. Our method precisely patterns multiple proteins in different spatial regions without gaps. We demonstrate the utility of our method by using this platform to analyze the crosstalk between platelet receptors, glycoprotein Ib and IIb–IIIa, in the context of platelet adhesion and signaling under flow. We show the clinical utility of this platform by applying it to analyze whole blood samples and to assess differences in the activation of platelets between healthy and diabetic patients.


 Please wait while we load your content...
Please wait while we load your content...