Research highlights: nanopore protein detection and analysis†
Abstract
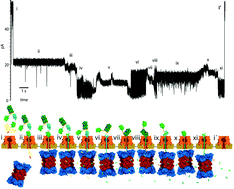
In this article we highlight recent work using nanopores to detect and study proteins. Nanopores are excellent single molecule sensors, capable of rapidly characterizing small molecules with relatively modest instrumentation requirements. Although the vast majority of recent effort and attention surrounding nanopores has centered on detection and sequencing of nucleic acids, proteins represent a more difficult and diverse analyte population, with a wide range of sizes, structures, charges, among other characteristics. Nanopores can be used to detect the presence of proteins of interest as well as to study their enzymatic activity, binding to ligands, and secondary structure. We highlight new work describing detection of specific protein species in solution by coupling them to a strand of carrier DNA that is used to electrophoretically transport the proteins through conical glass nanopores. Additionally, we spotlight another approach for nanopore detection of protein and other analytes through detection of their binding to aptamers—measurements which were quantitative to pM concentrations. Finally, we highlight studies in which protein secondary structure and folding energetics were studied through the use of an unfoldase enzyme coupled to a protein nanopore, a technique capable of detecting the effects of single amino acid mutations on the stability of the folded protein.

 Please wait while we load your content...
Please wait while we load your content...