Versatile multiple protein nanopatterning within a microfluidic channel for cell recruitment studies†
Abstract
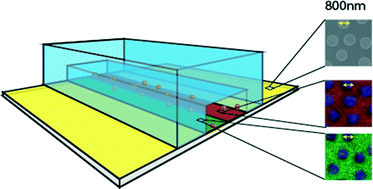
A novel approach combining self-assembly-based colloidal lithography and polydimethylsiloxane (PDMS) micromolding to generate complex protein nanopatterns for studying the mechanisms of leukocyte extravasation within microchannels is presented. Nanostructured surfaces sealed onto PDMS-molded microchannels are chemically functionalized in situ in an all-aqueous process to generate bi-functional chemical nanopatterns. Subsequent co-immobilization with proteins makes use of common non-covalent coupling (e.g. HIS-tags, FC-tags and biotin-tags), giving nanopatterns of arbitrary combinations of oriented, functional proteins. Up to three different proteins were simultaneously co-immobilized into the microchannel with nanoscale precision, demonstrating the complex patterns. As a proof-of-principle, a mimic of an inflamed endothelium was constructed using a macro- and nanoscale pattern of intercellular adhesion molecule 1 (ICAM1) and P-selectin, and the response of leukocytes through live cell imaging was measured. A clear result on the rolling behavior of the cells was observed with rolling limited to areas where ICAM1 and P-selectin are present. This micro/nano-interface will open new doors to investigations of how spatial distributions of proteins control cellular activity.

 Please wait while we load your content...
Please wait while we load your content...