High purity microfluidic sorting and analysis of circulating tumor cells: towards routine mutation detection†
Abstract
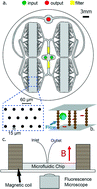
A new generation of the Ephesia cell capture technology optimized for CTC capture and genetic analysis is presented, characterized in depth and compared with the CellSearch system as a reference. This technology uses magnetic particles bearing tumour-cell specific EpCAM antibodies, self-assembled in a regular array in a microfluidic flow cell. 48 000 high aspect-ratio columns are generated using a magnetic field in a high throughput (>3 ml h−1) device and act as sieves to specifically capture the cells of interest through antibody–antigen interactions. Using this device optimized for CTC capture and analysis, we demonstrated the capture of epithelial cells with capture efficiency above 90% for concentrations as low as a few cells per ml. We showed the high specificity of capture with only 0.26% of non-epithelial cells captured for concentrations above 10 million cells per ml. We investigated the capture behavior of cells in the device, and correlated the cell attachment rate with the EpCAM expression on the cell membranes for six different cell lines. We developed and characterized a two-step blood processing method to allow for rapid processing of 10 ml blood tubes in less than 4 hours, and showed a capture rate of 70% for as low as 25 cells spiked in 10 ml blood tubes, with less than 100 contaminating hematopoietic cells. Using this device and procedure, we validated our system on patient samples using an automated cell immunostaining procedure and a semi-automated cell counting method. Our device captured CTCs in 75% of metastatic prostate cancer patients and 80% of metastatic breast cancer patients, and showed similar or better results than the CellSearch device in 10 out of 13 samples. Finally, we demonstrated the possibility of detecting cancer-related PIK3CA gene mutation in 20 cells captured in the chip with a good correlation between the cell count and the quantitation value Cq of the post-capture qPCR.

 Please wait while we load your content...
Please wait while we load your content...