Thermodynamics of force-dependent folding and unfolding of small protein and nucleic acid structures
Abstract
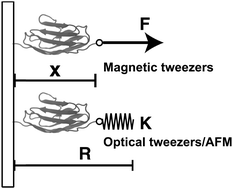
In this paper, we outline the theoretical framework for understanding the equilibrium force-dependent folding and unfolding transitions of protein domains and small nucleic acid structures, both having small rigid folded structures and highly flexible unfolded polymeric chain conformations. A complete statistical description of the state described by the probability function ρξ(n,x), is obtained, where n is an index denoting the structural state, and x is the extension of the molecule. ξ denotes an external constraint applied to the molecule, which is either a constant force or a harmonic spring attached to one end of the molecule. The extension probability distribution regardless of the structural state: 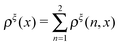 , the free energy landscape: −kBT ln(ρξ(x)), and the probability of the states regardless of the extension:
, the free energy landscape: −kBT ln(ρξ(x)), and the probability of the states regardless of the extension: 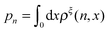 , are analyzed using the force-dependent structural transitions of the classic titin I27 domain as an example. The impact of different external constraints is also discussed.
, are analyzed using the force-dependent structural transitions of the classic titin I27 domain as an example. The impact of different external constraints is also discussed.
- This article is part of the themed collection: Mechanobiology

 Please wait while we load your content...
Please wait while we load your content...