Factors associated with elevated levels of antibiotic resistance genes in sewer sediments and wastewater†
Abstract
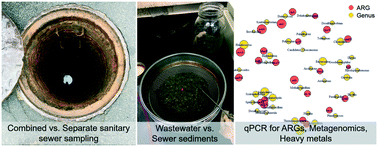
The sewer environment is a potential hotspot for the proliferation of antibiotic resistance genes (ARGs) and other hazardous microbial agents. Understanding the potential for ARG proliferation and retardation and/or accumulation in sewer sediments is of interest for protecting the health of sewage workers and the broader community in the event of sewer overflows as well as for interpreting sewage epidemiology data. To better understand this understudied environment for antibiotic resistance, a field survey was conducted to identify the factors that may control ARGs in sewer sediments and sewage. qPCR was performed for select ARGs and amplicon sequencing was performed for paired samples from combined and separate sanitary sewer systems. Metagenomic sequencing was performed on combined sewer sediments. The relative abundances of sul1, tet(O), tet(W), ermF, and vanA were higher in wastewater compared to sewer sediments, while NDM-1 was greater in sewer sediment and tet(G) was similar between the two matrices. NDM-1 was observed in sewer sediment but rarely above detection in wastewater in this study. This may indicate that larger/more frequent wastewater samples are needed for detection and/or that retardation and/or accumulation in sewage sediment may need to be considered when interpreting wastewater-based epidemiology data for ARGs. Random forest analyses indicated that season and conductivity were important variables and to a lesser extent so were pH, TSS, heavy metals, and sewer type for explaining the variance of the ARGs. These variables explained the 19–61% of the variance of sul1, tet(O), tet(G), and tet(W) quantified in wastewater. These variables performed less well for explaining the variance in sewer sediments (0.2–24%). Sewer sediment and wastewater had distinct microbial community structures and biomarkers for each are described. Metagenomics indicated that a high diversity of ARGs, including several of medical importance, was observed in the combined sewer sediment. This work provides insight into the complex sewer microbiome and the potential hazard posed by different sewer matrices.


 Please wait while we load your content...
Please wait while we load your content...
