DNA nuclease activity of Rev-coupled transition metal chelates†
Abstract
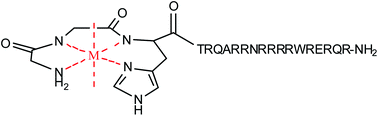
Artificial nucleases containing Rev-coupled metal chelates based on combinations of the transition metals Fe2+, Co2+, Ni2+, and Cu2+ and the chelators DOTA, DTPA, EDTA, NTA, tripeptide GGH, and tetrapeptide KGHK have been tested for DNA nuclease activity. Originally designed to target reactive transition metal chelates (M-chelates) to the HIV-1 Rev response element mRNA, attachment to the arginine-rich Rev peptide also increases DNA-binding affinity for the attached M-chelates. Apparent KD values ranging from 1.7 to 3.6 µM base pairs for binding of supercoiled pUC19 plasmid DNA by Ni–chelate–Rev complexes were observed, as a result of electrostatic attraction between the positively-charged Rev peptide and negatively-charged DNA. Attachment of M-chelates to the Rev peptide resulted in enhancements of DNA nuclease activity ranging from 1-fold (no enhancement) to at least 13-fold (for Cu–DTPA–Rev), for the rate of DNA nicking, with second order rate constants for conversion of DNAsupercoiled to DNAnicked up to 6 × 106 M−1 min−1, and for conversion of DNAnicked to DNAlinear up to 1 × 105 M−1 min−1. Freifelder–Trumbo analysis and the ratios of linearization and nicking rate constants (klin/knick) revealed concerted mechanisms for nicking and subsequent linearization of plasmid DNA for all of the Rev-coupled M-chelates, consistent with higher DNA residency times for the Rev-coupled M-chelates. Observed rates for Rev-coupled M-chelates were less skewed by differing DNA-binding affinities than for M-chelates lacking Rev, as a result of the narrow range of DNA-binding affinities observed, and therefore relationships between DNA nuclease activity and other catalyst properties, such as coordination unsaturation, the ability to consume ascorbic acid and generate diffusible radicals, and the identity of the metal center, are now clearly illustrated in light of the similar DNA-binding affinities of all M-chelate–Rev complexes. This work paints a clearer picture of the factors governing DNA nuclease activity by redox active M-chelates than was previously possible. The results demonstrate enhancement of DNA cleavage by use of a targeting sequence, but also clearly underscore that significant orientational factors are required for optimal reactivity at the metal center. Moreover, the studies confirm high selectivity for the target HIV RRE RNA at the most likely dosage concentrations, lending further support to the feasibility of designing and applying targeted catalytic metallodrugs.
- This article is part of the themed collection: Application of Inorganic Chemistry for non-Cancer Therapeutics

 Please wait while we load your content...
Please wait while we load your content...