Visualizing cellular machines with colocalization single molecule microscopy
Abstract
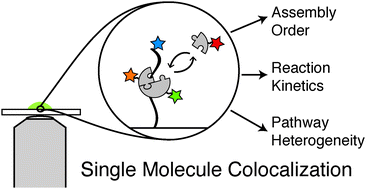
Many of the cell's macromolecular machines contain multiple components that transiently associate with one another. This compositional and dynamic complexity presents a challenge for understanding how these machines are constructed and function. Colocalization single molecule spectroscopy enables simultaneous observation of individual components of these machines in real-time and grants a unique window into processes that are typically obscured in ensemble assays. Colocalization experiments can yield valuable information about assembly pathways, compositional heterogeneity, and kinetics that together contribute to the development of richly detailed reaction mechanisms. This review focuses on recent advances in colocalization single molecule spectroscopy and how this technique has been applied to enhance our understanding of transcription, RNA splicing, and translation.
- This article is part of the themed collection: Single-molecule optical spectroscopy

 Please wait while we load your content...
Please wait while we load your content...