Proteotyping for the rapid identification of influenza virus and other biopathogens
Abstract
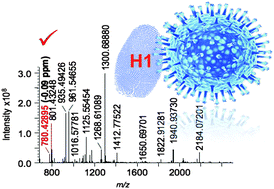
The influenza virus is one of the most deadly infectious agents known to man and has been responsible for the deaths of some hundred million lives throughout human history. The need to rapidly and reliably survey circulating virus strains down to the molecular level is ever present. This tutorial describes the development and application of a new proteotyping approach that harnesses the power of high resolution of
- This article is part of the themed collection: Chemical and biological detection

 Please wait while we load your content...
Please wait while we load your content...