Polymer models for the mechanisms of chromatin 3D folding: review and perspective
Abstract
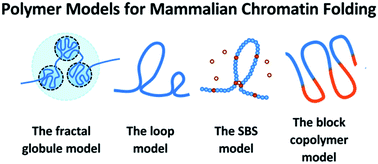
Recent experiments have provided unprecedented details on the hierarchical organization of the chromatin 3D structure and thus a great opportunity for understanding the mechanisms behind chromatin folding. As a bridge between experimental results and physical theory, coarse-grained polymer models of chromatin are of great value. Here, we review several popular models of chromatin folding, including the fractal globule model, loop models (the random loop model, the dynamic loop model, and the loop extrusion model), the string-and-binder switch model, and the block copolymer model. Physical models are still in great need to explain a larger variety of chromatin folding properties, especially structural features at different scales, their relation to the heterogeneous nature of the DNA sequence, and the highly dynamic nature of chromatin folding.
- This article is part of the themed collection: PCCP Perspectives


 Please wait while we load your content...
Please wait while we load your content...