Lysozyme–luteolin binding: molecular insights into the complexation process and the inhibitory effects of luteolin towards protein modification†
Abstract
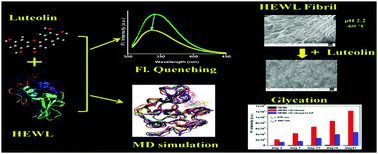
In the proposed work, the complexation of bioactive flavonoid luteolin with hen egg white lysozyme (HEWL) along with its inhibitory influence on HEWL modification has been explored with the help of multi-spectroscopic and computational methods. The binding affinity has been observed to be moderate in nature (in the order of 104 M−1) and the static quenching mechanism was found to be involved in the fluorescence quenching process. The binding constant (Kb) shows a progressive increase with the increase in temperature from (4.075 ± 0.046 × 104 M−1) at 293 K to (6.962 ± 0.024 × 104 M−1) at 313 K under experimental conditions. Spectroscopic measurements along with molecular docking calculations suggest that Trp62 is involved in the binding site of luteolin within the geometry of HEWL. The positive changes in enthalpy (ΔH = +19.99 ± 0.65 kJ mol−1) as well as entropy (ΔS = +156.28 ± 2.00 J K−1 mol−1) are indicative of the presence of hydrophobic forces that stabilize the HEWL–luteolin complex. The micro-environment around the Trp residues showed an increase in hydrophobicity as indicated by synchronous fluorescence (SFS), three dimensional fluorescence (3D) and red edge excitation (REES) studies. The % α-helix of HEWL showed a marked reduction upon binding with luteolin as indicated by circular dichroism (CD) and Fourier-transform infrared spectroscopy (FTIR) studies. Moreover, luteolin is situated at a distance of 4.275 ± 0.004 nm from the binding site as indicated by FRET theory, and the rate of energy transfer kET (0.063 ± 0.004 ns−1) has been observed to be faster than the donor decay rate (1/τD = 0.606 ns−1), which is indicative of the non-radiative energy transfer during complexation. Leaving aside the binding study, luteolin showed promising inhibitory effects towards the D-ribose mediated glycation of HEWL as well as towards HEWL fibrillation as studied by fluorescence emission and imaging studies. Excellent correlation with the experimental observations as well as precise location and dynamics of luteolin within the binding site has been obtained from molecular docking and molecular dynamics simulation studies.


 Please wait while we load your content...
Please wait while we load your content...