Entropy reduction in unfolded peptides (and proteins) due to conformational preferences of amino acid residues
Abstract
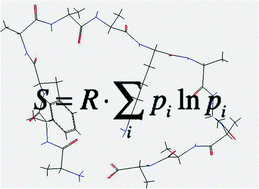
As established by several groups over the last 20 years, amino acid residues in unfolded peptides and proteins do not exhibit the unspecific random distribution as assumed by the classical random coil model. Individual amino acid residues in small peptides were found to exhibit different conformational preferences. Here, we utilize recently obtained conformational distributions of guest amino acid residues in GxG peptides to estimate their conformational entropy, which we find to be significantly lower than the entropy of an assumed random coil like distribution. Only at high temperature do backbone entropies approach random coil like values. We utilized the obtained backbone entropies of the investigated amino acid residues to estimate the loss of conformational entropy caused by a coil → helix transition and identified two subsets of amino acid residues for which the thus calculated entropy losses correlate well with the respective Gibbs energy of helix formation obtained for alanine based host–guest systems. Calculated and experimentally derived entropic losses were found to be in good agreement. For most of the amino acid residues investigated entropic losses derived from our GxG distributions correlate very well with corresponding values recently obtained from MD simulations biased by conformational propensities derived from truncated coil libraries. Both, conformational entropy and the entropy of solvation exhibit a strong, residue specific temperature dependence, which can be expected to substantially affect the stability of unfolded states. Altogether, our results provide strong evidence for the notion that conformational preferences of amino acid residues matter with regard to the thermodynamics of peptide and protein folding.

 Please wait while we load your content...
Please wait while we load your content...