Understanding the interaction of DNA–RNA nucleobases with different ZnO nanomaterials†
Abstract
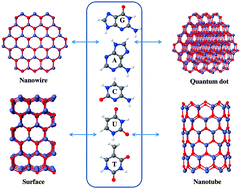
Due to the potential application of different nanostructure materials in biomedical nanotechnologies, understanding the interaction between the inorganic nanoparticles and biological molecules at the atomic level is of paramount importance. We present here the results of our theoretical investigation of the interaction of different nucleotide bases – adenine (A), guanine (G), cytosine (C), thymine (T) and uracil (U) of deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) – with different ZnO nanoparticles, such as ZnO nanowires (NWs), nanotubes (NTs), surfaces and quantum dots (QDs). As the size of the systems we studied is relatively large, we have used the self-consistent-charge density-functional tight-binding (SCC-DFTB) method to optimize the complex systems. We have studied in detail the site-specific binding nature and the adsorption strength of these nucleobases with different ZnO nanoparticles. The calculated binding energy order and the interaction strength of nucleobases are very much dependent on the nature of the nanoparticle surfaces and are different for different nanostructures. In most of the cases ZnO prefers to bind either through the top site of the nucleobases or with the ring nitrogen atom having a lone pair relative to other binding sites of the bases.

 Please wait while we load your content...
Please wait while we load your content...