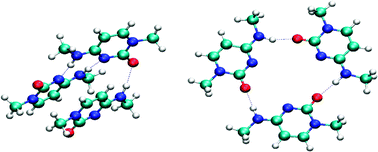
A new database of nucleic acid base trimers has been developed that includes 141 geometries and stabilization energies obtained at the RI-MP2 level of theory with the TZVPP basis set. Compared to previously compiled biologically oriented databases, this new construct includes considerably more complicated structures; the various intermolecular interactions in the trimers are quite heterogeneous and in particular include simultaneous hydrogen bonding and stacking interactions, which is similar to the situation in actual biopolymers. Validation against these benchmark data is therefore a more demanding task for approximate models, since correct descriptions of all energy terms are unlikely to be accomplished by fortuitous cancellations of systematic errors. The density functionals TPSS (both with and without an empirical dispersion term), PWB6K, M05-2X, and BH&H, and the self-consistent charge density functional tight binding method augmented with an empirical dispersion term (SCC-DFTB-D) were assessed for their abilities accurately to compute structures and energies. The best reproduction of the BSSE corrected RI-MP2 stabilization energies was achieved by the TPSS functional (TZVPP basis set) combined with empirical dispersion; removal of the dispersion correction leads to significantly degraded performance. The M05-2X and PWB6K functionals performed very well in reproducing the RI-MP2 geometries, but showed a systematic moderate underestimation of the magnitude of base stacking interactions. The SCC-DFTB-D method predicts geometries in fair agreement with RI-MP2; given its computational efficiency it represents a good option for initial scanning of analogous biopolymeric potential energy surfaces. BH&H gives geometries of comparable quality to the other functionals but significantly overestimates interaction energies other than stacking.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...