High-throughput screening of triplex DNA binders from complicated samples by 96-well pate format in conjunction with peak area-fading UHPLC-Orbitrap MS†
Abstract
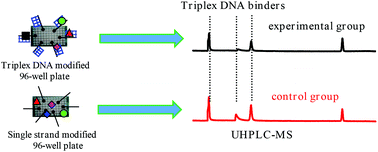
Conventional strategies for the screening of DNA triplex binders cannot be used for complicated samples, such as ligand libraries created by combinatorial chemistry or from natural product extracts. In the current study, an ultra-high-performance liquid chromatography coupled with an Orbitrap mass spectrometry (UHPLC-Orbitrap-MS)-based approach, which we call peak area-fading (PAF) UHPLC-Orbitrap-MS and was designed for just such a purpose, is reported. The triplex DNA modified 96-well plate and the single stranded oligonucleotide modified 96-well plate (as control) were incubated with ligand libraries, and the unbound ligands were directly determined via UHPLC-ESI-MS. The binders were detected through the decrease (fading) in the peak areas compared to those of the control group. Several factors, such as incubation time, incubation temperature, and buffer, which might affect the binding affinity and reproducibility, were optimized. The potential of the approach was examined using the extracts of Rhizoma Coptidis and Phellodendron chinense Schneid cortexe. The triplex DNA-binding capabilities of the five components (epiberberine, coptisine, jatrorrhizine, berberrubine, and columbamine) were found for the first time, indicating their efficiency for the analysis of complicated samples. In contrast to our previous study, which suffered from a serious drawback of poor reproducibility, this method is more robust and more suitable for high-throughput measurements, opening a new experimental strategy in assessing large libraries of potential drug candidates that work by forming a drug/DNA complex.


 Please wait while we load your content...
Please wait while we load your content...