Energy-resolved collision-induced dissociation pathways of model N-linked glycopeptides: implications for capturing glycan connectivity and peptide sequence in a single experiment†
Abstract
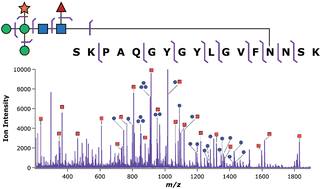
Tandem mass spectrometry (MS/MS) of glycopeptides stands among the principal analytical approaches for assessing protein glycosylation in a site-specific manner. The aims of such experiments are often to determine the monosaccharide connectivity of the glycan, the amino acid sequence of the peptide, and the site of glycan attachment. This level of detail is often difficult to achieve using any single ion dissociation method; however, precedent does exist for use of collision-induced dissociation (CID) to establish either the connectivity of the oligosaccharide or the sequence of the polypeptide depending upon the applied collision energy. Unfortunately, the relative energy requirements for glycan and peptide cleavage have not been thoroughly characterized with respect to specific physicochemical characteristics of the precursor ions. This report describes case studies on the energy-resolved CID pathways of model tryptic glycopeptides derived from Erythrina cristagalli lectin and bovine ribonuclease B. While glycopeptide ions having disparate physical and chemical characteristics shared strikingly similar qualitative responses to increasing vibrational energy deposition, the absolute collision energies at which either glycan or peptide fragmentations were accessed varied substantially among the precursor ions examined. Nevertheless, these data suggest that the energy requirements for peptide and glycan cleavage may be somewhat predictable based on characteristics of the precursor ion. The practical usefulness of these observations was demonstrated through implementation of online collision energy modulation such that both glycan and peptide fragmentation were captured in the same spectrum, providing near-exhaustive glycopeptide characterization in a single experiment. Overall, these results highlight the potential to further extend the capabilities of CID in the context of glycoproteomics.

 Please wait while we load your content...
Please wait while we load your content...