CarsiDock: a deep learning paradigm for accurate protein–ligand docking and screening based on large-scale pre-training†
Abstract
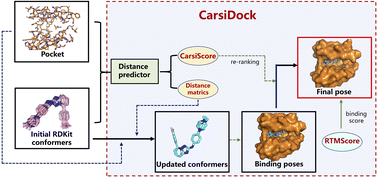
The expertise accumulated in deep neural network-based structure prediction has been widely transferred to the field of protein–ligand binding pose prediction, thus leading to the emergence of a variety of deep learning-guided docking models for predicting protein–ligand binding poses without relying on heavy sampling. However, their prediction accuracy and applicability are still far from satisfactory, partially due to the lack of protein–ligand binding complex data. To this end, we create a large-scale complex dataset containing ∼9 M protein–ligand docking complexes for pre-training, and propose CarsiDock, the first deep learning-guided docking approach that leverages pre-training of millions of predicted protein–ligand complexes. CarsiDock contains two main stages, i.e., a deep learning model for the prediction of protein–ligand atomic distance matrices, and a translation, rotation and torsion-guided geometry optimization procedure to reconstruct the matrices into a credible binding pose. The pre-training and multiple innovative architectural designs facilitate the dramatically improved docking accuracy of our approach over the baselines in terms of multiple docking scenarios, thereby contributing to its outstanding early recognition performance in several retrospective virtual screening campaigns. Further explorations demonstrate that CarsiDock can not only guarantee the topological reliability of the binding poses but also successfully reproduce the crucial interactions in crystalized structures, highlighting its superior applicability.
- This article is part of the themed collection: Most popular 2024 physical, theoretical and computational chemistry articles


 Please wait while we load your content...
Please wait while we load your content...