Removal of hemolysis interference in serum Raman spectroscopy by multivariate curve resolution analysis for accurate classification of oral cancers†
Abstract
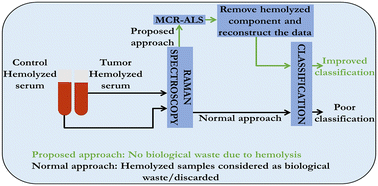
Serum is an important biofluid that accurately reflects pathology and is used for early diagnosis of a wide range of diseases. Like most serological assays, Raman spectroscopic measurements also require serum samples free of hemolysis, i.e., breakdown of red blood cells, as their strong contribution to the Raman spectrum can mask subtle biomolecular changes in serum leading to inaccurate results. Therefore, hemolyzed samples are routinely rejected for serum Raman studies. However, in most cases, it would be difficult to get fresh samples, thereby reducing the sample size for chemometric analysis. In this work, we employed multivariate curve resolution-alternating least squares (MCR-ALS) analysis to extract pure biomolecular components and tried to digitally remove interference due to hemolysis in serum Raman spectroscopy. We demonstrate its application using hemolyzed/non-hemolyzed serum from control hamsters (untreated) and hamsters with 7,12-dimethylbenz[a]anthracene (DMBA)-induced oral tumors. Our results clearly show that even Raman spectra of hemolyzed serum samples can be pre-processed and used to make a differential diagnosis with high accuracy. We believe that the proposed method has huge potential in Raman diagnostics as it can be employed for the so-called routinely used non-hemolyzed serum samples to further improve diagnostic accuracies as shown in this work.


 Please wait while we load your content...
Please wait while we load your content...