MetalProGNet: a structure-based deep graph model for metalloprotein–ligand interaction predictions†
Abstract
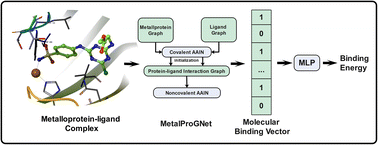
Metalloproteins play indispensable roles in various biological processes ranging from reaction catalysis to free radical scavenging, and they are also pertinent to numerous pathologies including cancer, HIV infection, neurodegeneration, and inflammation. Discovery of high-affinity ligands for metalloproteins powers the treatment of these pathologies. Extensive efforts have been made to develop in silico approaches, such as molecular docking and machine learning (ML)-based models, for fast identification of ligands binding to heterogeneous proteins, but few of them have exclusively concentrated on metalloproteins. In this study, we first compiled the largest metalloprotein–ligand complex dataset containing 3079 high-quality structures, and systematically evaluated the scoring and docking powers of three competitive docking tools (i.e., PLANTS, AutoDock Vina and Glide SP) for metalloproteins. Then, a structure-based deep graph model called MetalProGNet was developed to predict metalloprotein–ligand interactions. In the model, the coordination interactions between metal ions and protein atoms and the interactions between metal ions and ligand atoms were explicitly modelled through graph convolution. The binding features were then predicted by the informative molecular binding vector learned from a noncovalent atom–atom interaction network. The evaluation on the internal metalloprotein test set, the independent ChEMBL dataset towards 22 different metalloproteins and the virtual screening dataset indicated that MetalProGNet outperformed various baselines. Finally, a noncovalent atom–atom interaction masking technique was employed to interpret MetalProGNet, and the learned knowledge accords with our understanding of physics.


 Please wait while we load your content...
Please wait while we load your content...