Building Streptomyces albus as a chassis for synthesis of bacterial terpenoids†
Abstract
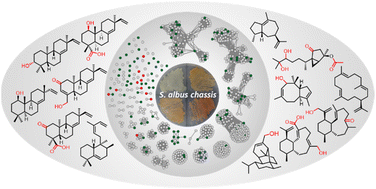
Terpenoids comprise the most chemically and structurally diverse family of natural products. In contrast to the huge numbers of terpenoids discovered from plants and fungi, only a relatively small number of terpenoids were reported from bacteria. Recent genomic data in bacteria suggest that a large number of biosynthetic gene clusters encoding terpenoids remain uncharacterized. In order to enable the functional characterization of terpene synthase and relevant tailoring enzymes, we selected and optimized an expression system based on a Streptomyces chassis. Through genome mining, 16 distinct bacterial terpene biosynthetic gene clusters were selected and 13 of them were successfully expressed in the Streptomyces chassis, leading to characterization of 11 terpene skeletons including three new ones, representing an ∼80% success rate. In addition, after functional expression of tailoring genes, 18 novel distinct terpenoids were isolated and characterized. This work demonstrates the advantages of a Streptomyces chassis which not only enabled the successful production of bacterial terpene synthases, but also enabled functional expression of tailoring genes, especially P450, for terpenoid modification.


 Please wait while we load your content...
Please wait while we load your content...