Sensing platform for nucleic-acid detection based on a 2-aminopurine probe sheared by trans-cleavage activity of the CRISPR/Cas12a system†
Abstract
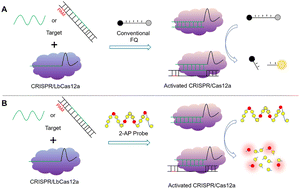
Target double-stranded DNA (dsDNA) or single-stranded DNA (ssDNA) can activate the trans-cleavage activity of the CRISPR/Cas12a, cutting the surrounding non-target ssDNA arbitrarily. In a typical CRISPR/Cas12a system, this non-target ssDNA, with a fluorescent tag and its quencher incorporated at both ends (ssDNA-FQ), is usually used as the reporter. Here, a 2-aminopurine probe (T-pro 4), made by inserting four 2-APs in non-target ssDNA, was screened for using as a reporter in the CRISPR/Cas12a system. Compared with ssDNA-FQ, each 2-AP probe is cleaved by the activated CRISPR/Cas12a system, multi-unit signals are generated. Therefore, the CRISPR/Cas12a system using the 2-AP probe as a reporter may be more sensitive than the CRISPR/Cas12a system which uses ssDNA-FQ as the reporter. We achieved ssDNA detection at as little as 10−11 M using the 2-AP probe as the reporter in the CRISPR/Cas12a system. Compared to the CRISPR/Cas12a system using ssDNA-FQ as the reporter, its sensitivity increased by an order of magnitude. Furthermore, the method that combines PCR and the 2-AP-probe-mediated CRISPR/Cas12a system can detect goat pox virus (GTPV) down to 8.35 × 10−2 copies per μL, 10 times lower than the method that combines PCR and the ssDNA-FQ-mediated CRISPR/Cas12a system. These results indicate that the CRISPR/Cas12a system using the screened 2-AP probe as a reporter has potential in highly sensitive detection of viruses.
- This article is part of the themed collection: Analyst HOT Articles 2023


 Please wait while we load your content...
Please wait while we load your content...