Identification of two-dimensional copper signatures in human blood for bladder cancer with machine learning†
Abstract
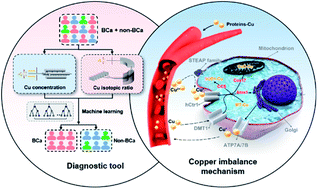
Currently, almost all available cancer biomarkers are based on concentrations of compounds, often suffering from low sensitivity, poor specificity, and false positive or negative results. The stable isotopic composition of elements provides a different dimension from the concentration and has been widely used as a tracer in geochemistry. In health research, stable isotopic analysis has also shown potential as a new diagnostic/prognostic tool, which is still in the nascent stage. Here we discovered that bladder cancer (BCa) could induce a significant variation in the ratio of natural copper isotopes (65Cu/63Cu) in the blood of patients relative to benign and healthy controls. Such inherent copper isotopic signatures permitted new insights into molecular mechanisms of copper imbalance underlying the carcinogenic process. More importantly, to enhance the diagnostic capability, a machine learning model was developed to classify BCa and non-BCa subjects based on two-dimensional copper signatures (copper isotopic composition and concentration in plasma and red blood cells) with a high sensitivity, high true negative rate, and low false positive rate. Our results demonstrated the promise of blood copper signatures combined with machine learning as a versatile tool for cancer research and potential clinical application.


 Please wait while we load your content...
Please wait while we load your content...