Multi-omic profiling of histone variant H3.3 lysine 27 methylation reveals a distinct role from canonical H3 in stem cell differentiation†
Abstract
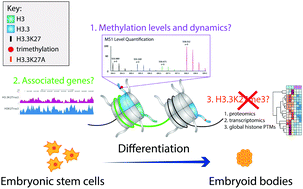
Histone variants, such as histone H3.3, replace canonical histones within the nucleosome to alter chromatin accessibility and gene expression. Although the biological roles of selected histone post-translational modifications (PTMs) have been extensively characterized, the potential differences in the function of a given PTM on different histone variants is almost always elusive. By applying proteomics and genomics techniques, we investigate the role of lysine 27 tri-methylation specifically on the histone variant H3.3 (H3.3K27me3) in the context of mouse embryonic stem cell pluripotency and differentiation as a model system for development. We demonstrate that while the steady state overall levels of methylation on both H3K27 and H3.3K27 decrease during differentiation, methylation dynamics studies indicate that methylation on H3.3K27 is maintained more than on H3K27. Using a custom-made antibody, we identify a unique enrichment of H3.3K27me3 at lineage-specific genes, such as olfactory receptor genes, and at binding motifs for the transcription factors FOXJ2/3. REST, a predicted FOXJ2/3 target that acts as a transcriptional repressor of terminal neuronal genes, was identified with H3.3K27me3 at its promoter region. H3.3K27A mutant cells confirmed an upregulation of FOXJ2/3 targets upon the loss of methylation at H3.3K27. Thus, while canonical H3K27me3 has been characterized to regulate the expression of transcription factors that play a general role in differentiation, our work suggests H3.3K27me3 is essential for regulating distinct terminal differentiation genes. This work highlights the importance of understanding the effects of PTMs not only on canonical histones but also on specific histone variants, as they may exhibit distinct roles.


 Please wait while we load your content...
Please wait while we load your content...