The impact of AlphaFold2 on experimental structure solution
Abstract
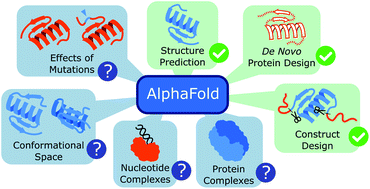
AlphaFold2 is a machine-learning based program that predicts a protein structure based on the amino acid sequence. In this article, we report on the current usages of this new tool and give examples from our work in the Coronavirus Structural Task Force. With its unprecedented accuracy, it can be utilized for the design of expression constructs, de novo protein design and the interpretation of Cryo-EM data with an atomic model. However, these methods are limited by their training data and are of limited use to predict conformational variability and fold flexibility; they also lack co-factors, post-translational modifications and multimeric complexes with oligonucleotides. They also are not always perfect in terms of chemical geometry. Nevertheless, machine learning-based fold prediction is a game changer for structural bioinformatics and experimentalists alike, with exciting developments ahead.
- This article is part of the themed collections: Computational protein design and structure prediction: Celebrating the 2024 Nobel Prize in Chemistry and Challenges in biological cryo electron microscopy


 Please wait while we load your content...
Please wait while we load your content...