BRD4: quantum mechanical protein–ligand binding free energies using the full-protein DFT-based QM-PBSA method†
Abstract
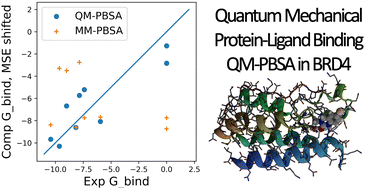
Fully quantum mechanical approaches to calculating protein–ligand free energies of binding have the potential to reduce empiricism and explicitly account for all physical interactions responsible for protein–ligand binding. In this study, we show a realistic test of the linear-scaling DFT-based QM-PBSA method to estimate quantum mechanical protein–ligand binding free energies for a set of ligands binding to the pharmaceutical drug-target bromodomain containing protein 4 (BRD4). We show that quantum mechanical QM-PBSA is a significant improvement over traditional MM-PBSA in terms of accuracy against experiment and ligand rank ordering and that the quantum and classical binding energies are converged to a similar degree. We test the interaction entropy and normal mode entropy correction terms to QM- and MM-PBSA.


 Please wait while we load your content...
Please wait while we load your content...