Structure and diffusive dynamics of aspartate α-decarboxylase (ADC) liganded with d-serine in aqueous solution†
Abstract
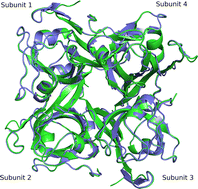
Incoherent neutron spectroscopy, in combination with dynamic light scattering, was used to investigate the effect of ligand binding on the center-of-mass self-diffusion and internal diffusive dynamics of Escherichia coli aspartate α-decarboxylase (ADC). The X-ray crystal structure of ADC in complex with the D-serine inhibitor was also determined, and molecular dynamics simulations were used to further probe the structural rearrangements that occur as a result of ligand binding. These experiments reveal that D-serine forms hydrogen bonds with some of the active site residues, that higher order oligomers of the ADC tetramer exist on ns–ms time-scales, and also show that ligand binding both affects the ADC internal diffusive dynamics and appears to further increase the size of the higher order oligomers.


 Please wait while we load your content...
Please wait while we load your content...