Listeria monocytogenes utilizes the ClpP1/2 proteolytic machinery for fine-tuned substrate degradation at elevated temperatures†
Abstract
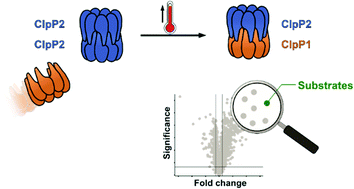
Listeria monocytogenes exhibits two ClpP isoforms (ClpP1/ClpP2) which assemble into a heterooligomeric complex with enhanced proteolytic activity. Herein, we demonstrate that the formation of this complex depends on temperature and reaches a maximum ratio of about 1 : 1 at 30 °C, while almost no complex formation occurred below 4 °C. In order to decipher the role of the two isoforms at elevated temperatures, we constructed L. monocytogenes ClpP1, ClpP2 and ClpP1/2 knockout strains and analyzed their protein regulation in comparison to the wild type (WT) strain via whole proteome mass-spectrometry (MS) at 37 °C and 42 °C. While the ΔclpP1 strain only altered the expression of very few proteins, the ΔclpP2 and ΔclpP1/2 strains revealed the dysregulation of many proteins at both temperatures. These effects were corroborated by crosslinking co-immunoprecipitation MS analysis. Thus, while ClpP1 serves as a mere enhancer of protein degradation in the heterocomplex, ClpP2 is essential for ClpX binding and functions as a gatekeeper for substrate entry. Applying an integrated proteomic approach combining whole proteome and co-immunoprecipitation datasets, several putative ClpP2 substrates were identified in the context of different temperatures and discussed with regards to their function in cellular pathways such as the SOS response.


 Please wait while we load your content...
Please wait while we load your content...