IVT cell-free biosensors for tetracycline and macrolide detection based on allosteric transcription factors (aTFs)†
Abstract
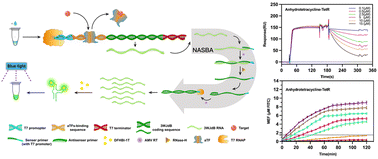
In recent years, the issue of food safety has received a lot of attention. The Food and Drug Administration (FDA) prescribes the antibiotic's maximum residue limit (MRL) in food production. The standard detection methods of antibiotics are liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) and high-performance liquid chromatography (HPLC), with complex operations and precision instruments. In this study, allosteric transcription factor (aTF)-based in vitro transcription (IVT) cell-free biosensors were developed for tetracyclines and macrolides with nucleic acid sequence-based amplification (NASBA). Characterization of binding and dissociation processes between aTF and DNA was carried out by BIAcore assay and electrophoretic mobility shift assay (EMSA). BIAcore was innovatively used to directly observe the real-time process of binding and dissociation of aTF with DNA. The biosensors produce more fluorescence RNA when target antibiotics are added to the three-way junction dimeric Broccoli (3WJdB). Four tetracyclines and two macrolides were quantified in the 0.5–15 μM range, while erythromycin and clarithromycin were detected over a range of 0.1–15 μM. NASBA, commonly used for viral detection, was used to amplify 3WJdB RNA generated by IVT, which greatly increased the LOD for tetracyclines and macrolides to 0.01 μM. The use of biosensors in milk samples demonstrated their on-site detection performance. Overall, our proposed biosensors are simple, rapid, selective, and sensitive, with the potential for field application.


 Please wait while we load your content...
Please wait while we load your content...