Development of novel enzyme immobilization methods employing formaldehyde or triethoxysilylbutyraldehyde to fabricate immobilized enzyme microreactors for peptide mapping†
Abstract
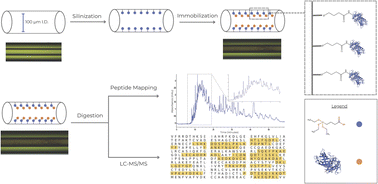
The digestion of proteins with proteolytic enzymes has expedited the analysis of peptide mapping. Here, we compared the digestion efficiency of soluble chymotrypsin (CT) with two immobilized CT preparations using bovine serum albumin (BSA) as the substrate. An efficient method of immobilizing chymotrypsin using formaldehyde (FA) was optimized and the conditions were applied to assess a novel immobilization reagent, triethoxysilylbutaraldehyde (TESB). Efforts to determine the best enzyme-to-substrate (E : S) ratios during digestion of denatured BSA with single-use FA–CT enzyme particles were performed by adjusting the amount of substrate used. An E : S ratio of 10 : 1 was found to be best based on the LC-MS/MS analysis data showing sequence coverage of 67%. Fabrication of immobilized enzyme microreactors (IMERs) was carried out using both (3-aminopropyl)triethoxysilane (APTES) with the idealized conditions with FA, as well as the novel procedure utilizing TESB for a proof of concept open-tubular IMER. It was found that the FA-APTES IMER had a sequence coverage of 6%, while the TESB IMER had 29% sequence coverage from MS analysis. The application of TESB in enzyme immobilization has the potential to facilitate a greater degree of enzymatic digestion with higher sequence coverage than traditional immobilization or crosslinking reagents for bottom-up proteomics.


 Please wait while we load your content...
Please wait while we load your content...