Direct decarboxylation of ten-eleven translocation-produced 5-carboxylcytosine in mammalian genomes forms a new mechanism for active DNA demethylation†
Abstract
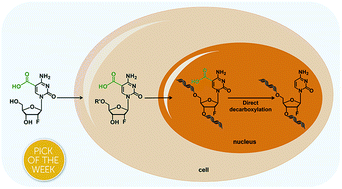
DNA cytosine methylation (5-methylcytosine, 5mC) is the most important epigenetic mark in higher eukaryotes. 5mC in genomes is dynamically controlled by writers and erasers. DNA (cytosine-5)-methyltransferases (DNMTs) are responsible for the generation and maintenance of 5mC in genomes. Active demethylation of 5-methylcytosine (5mC) is achieved by ten-eleven translocation (TET) dioxygenase-mediated oxidation of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). 5fC and 5caC are further processed by thymine DNA glycosylase (TDG)-initiated base excision repair (BER) to restore unmodified cytosines. The TET-TDG-BER pathway could cause the production of DNA strand breaks and therefore jeopardize the integrity of genomes. Here, we investigated the direct decarboxylation of 5caC in mammalian genomes by using metabolic labeling with 2′-fluorinated 5caC (F-5caC) and mass spectrometry analysis. Our results clearly demonstrated the decarboxylation of 5caC occurring in mammalian genomes, which unveiled that, in addition to the TET-TDG-BER pathway, the direct decarboxylation of TET-produced 5caC constituted a new pathway for active demethylation of 5mC in mammalian genomes.
- This article is part of the themed collection: 2021 ChemSci Pick of the Week Collection


 Please wait while we load your content...
Please wait while we load your content...