Enhanced sampling molecular dynamics simulations correctly predict the diverse activities of a series of stiff-stilbene G-quadruplex DNA ligands†
Abstract
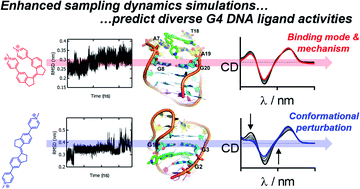
Ligands with the capability to bind G-quadruplexes (G4s) specifically, and to control G4 structure and behaviour, offer great potential in the development of novel therapies, technologies and functional materials. Most known ligands bind to a pre-formed topology, but G4s are highly dynamic and a small number of ligands have been discovered that influence these folding equilibria. Such ligands may be useful as probes to understand the dynamic nature of G4 in vivo, or to exploit the polymorphism of G4 in the development of molecular devices. To date, these fascinating molecules have been discovered serendipitously. There is a need for tools to predict such effects to drive ligand design and development, and for molecular-level understanding of ligand binding mechanisms and associated topological perturbation of G4 structures. Here we study the G4 binding mechanisms of a family of stiff-stilbene G4 ligands to human telomeric DNA using molecular dynamics (MD) and enhanced sampling (metadynamics) MD simulations. The simulations predict a variety of binding mechanisms and effects on G4 structure for the different ligands in the series. In parallel, we characterize the binding of the ligands to the G4 target experimentally using NMR and CD spectroscopy. The results show good agreement between the simulated and experimentally observed binding modes, binding affinities and ligand-induced perturbation of the G4 structure. The simulations correctly predict ligands that perturb G4 topology. Metadynamics simulations are shown to be a powerful tool to aid development of molecules to influence G4 structure, both in interpreting experiments and to help in the design of these chemotypes.


 Please wait while we load your content...
Please wait while we load your content...